scphylo.tl.rf#
- scphylo.tl.rf(df_grnd, df_sol)[source]#
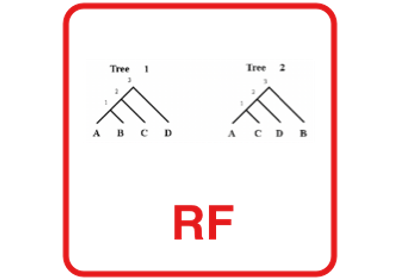
Robinson-Foulds score.
The Robinson–Foulds or symmetric difference metric is defined as (A + B) where A is the number of partitions of data implied by the first tree but not the second tree and B is the number of partitions of data implied by the second tree but not the first tree (although some software implementations divide the RF metric by 2 and others scale the RF distance to have a maximum value of 1).
- Parameters
df_grnd¶ (
pandas.DataFrame) – The first genotype matrix (e.g. ground truth) This matrix must be conflict-free.df_sol¶ (
pandas.DataFrame) – The second genotype matrix (e.g. solution/inferred) This matrix must be conflict-free.
- Returns
Similarity out of one.
- Return type